10 NEBNext versus scCUTseq
This sections produces all the figures used in Supplementary Figure 8.
# Source setup file
source("./functions/setup.R")
# Load functions
source("./functions/plotHeatmap.R")
source("./functions/plotProfile.R")10.1 Validating detected deletions using bulk NEBNext sequencing
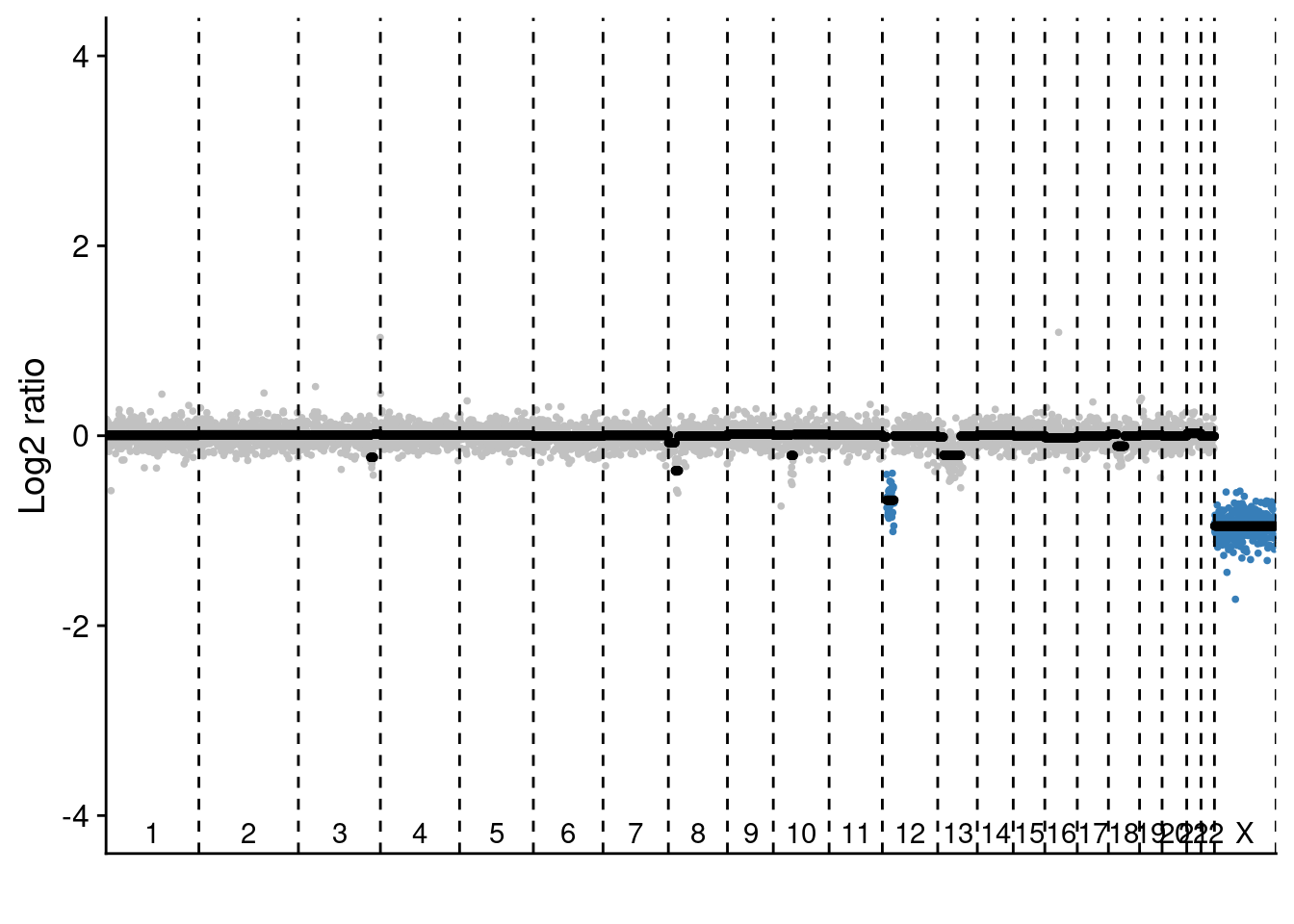
We have also performed bulk NEBNext sequencing of two sections in P6 where we saw a high number of pseudodiploid cells.
# Load in data
sccutseq = readRDS("./data/P6_cnv.rds")
annotation = fread("./annotation/P6.tsv", header = FALSE)
# The NEBNext sequencing we show is performed on section L2C2 and L3C3
libraries_sccut = annotation[V2 %in% c("L2C2", "L3C3"), V1]
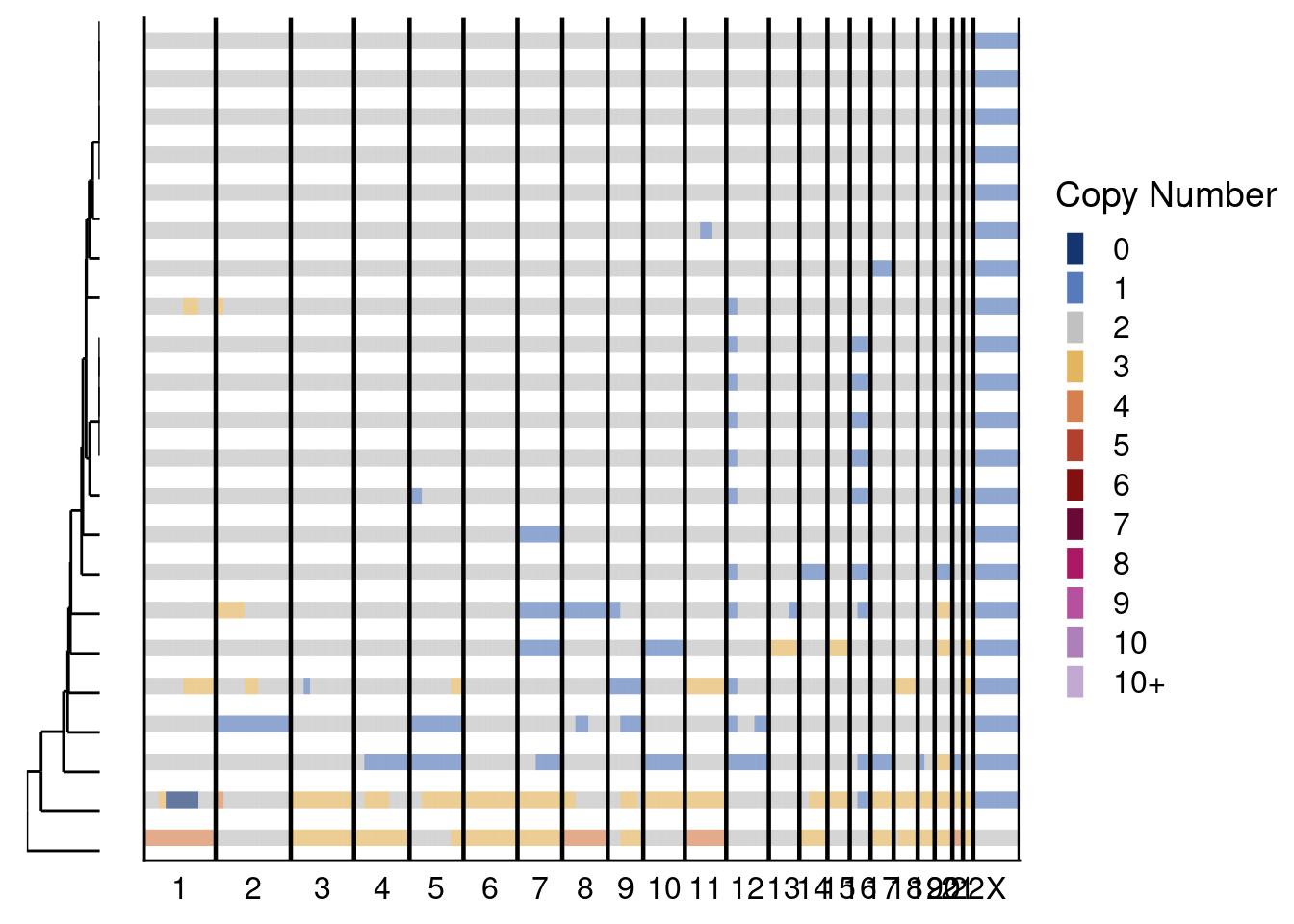
# First plot genomewide heatmaps.
sccutseq_profiles = sccutseq$copynumber[, sccutseq$stats[classifier_prediction == "good", sample], with = FALSE]
sccutseq_profiles_l2c2 = sccutseq_profiles[, grepl(libraries_sccut[1], colnames(sccutseq_profiles)), with = F]
sccutseq_profiles_l3c3 = sccutseq_profiles[, grepl(libraries_sccut[2], colnames(sccutseq_profiles)), with = F]
# Plot heatmaps
# L2C3
plotHeatmap(sccutseq_profiles_l2c2, sccutseq$bins, linesize = 3)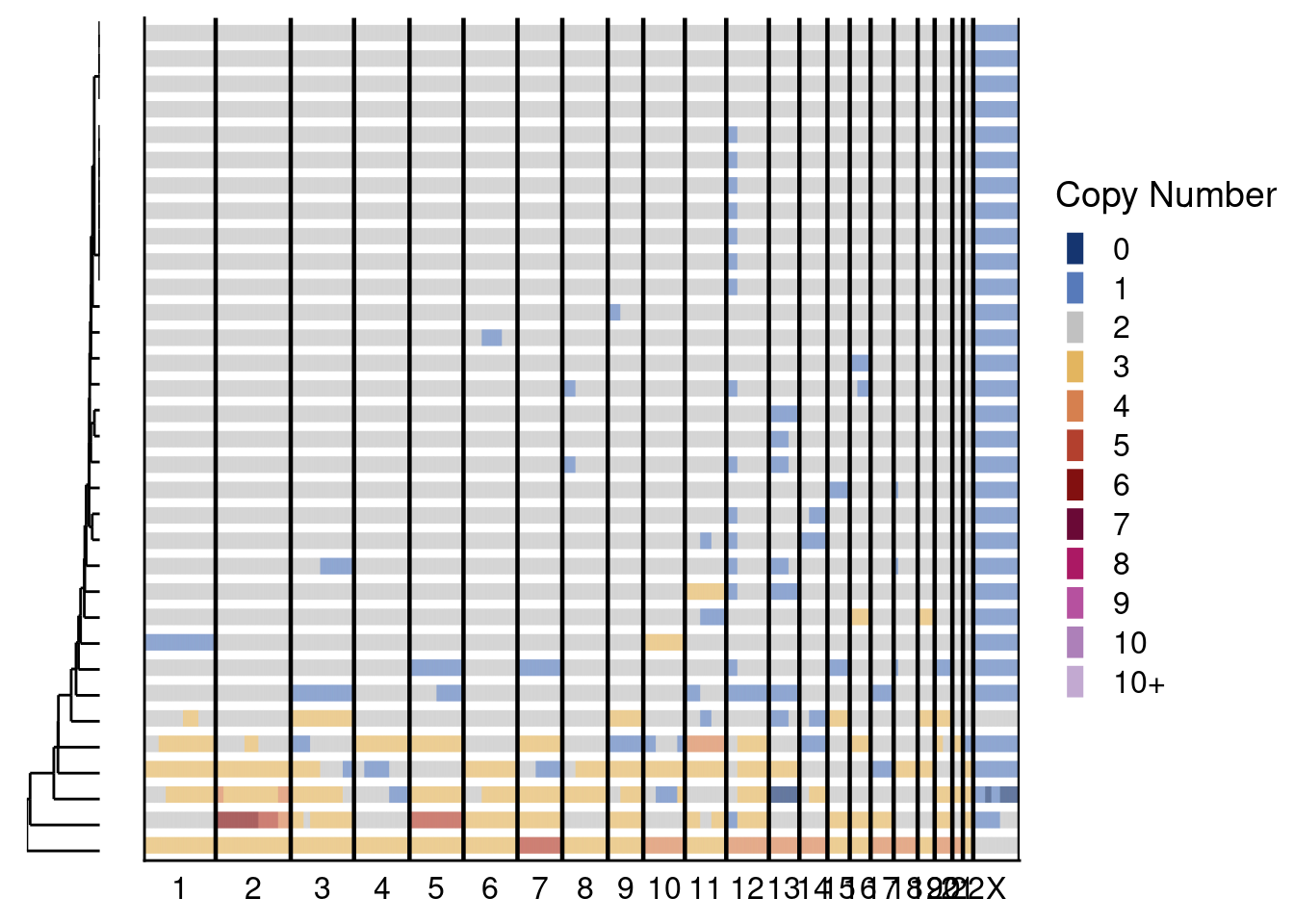
# Load NEB data
NEB = readRDS("./data/BC282_cnv.rds")
# Select samples
samples = c("NZ279", "NZ280") #NZ279 is L2C2 and NZ280 is L3C3
# Plot profiles
# L2C2
plotProfile(NEB$segments[[samples[1]]], NEB$counts_lrr[[samples[[1]]]], bins = NEB$bins, sc = FALSE)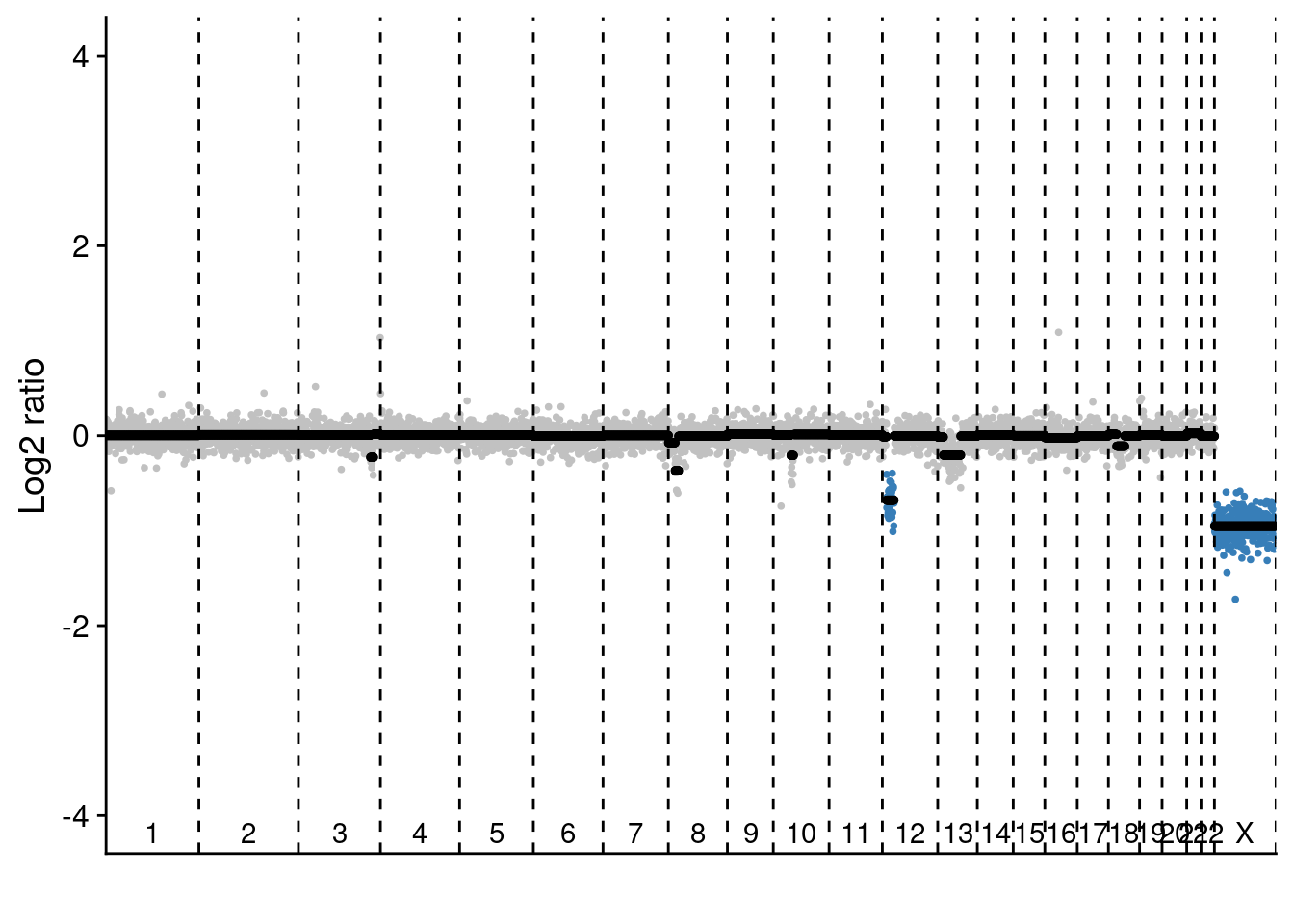
# L3C3
plotProfile(NEB$segments[[samples[1]]], NEB$counts_lrr[[samples[[1]]]], bins = NEB$bins, sc = FALSE)