8 Copy number profiles of brain neurons and skeletal muscle cells
This sections produces all the figures used in Supplementary Figure 6.
# Source setup file
source("./functions/setup.R")
# Load functions
source("./functions/plotHeatmap.R")8.1 Plot genomewide heatmaps
First load data and then plot genomewide data using the plotHeatmap() function
# Load all profiles
dg20 = readRDS("./data/DG20_cnv.rds")
dg21 = readRDS("./data/DG21_cnv.rds")
dg22 = readRDS("./data/DG22_cnv.rds")
dg33 = readRDS("./data/DG33_cnv.rds")
dg39 = readRDS("./data/DG39_cnv.rds")
dg40 = readRDS("./data/DG40_cnv.rds")
# Go through each library and select HQ cells
dg20_cn = dg20$copynumber[, dg20$stats[classifier_prediction == "good", sample], with = FALSE]
dg21_cn = dg21$copynumber[, dg21$stats[classifier_prediction == "good", sample], with = FALSE]
dg22_cn = dg22$copynumber[, dg22$stats[classifier_prediction == "good", sample], with = FALSE]
dg33_cn = dg33$copynumber[, dg33$stats[classifier_prediction == "good", sample], with = FALSE]
dg39_cn = dg39$copynumber[, dg39$stats[classifier_prediction == "good", sample], with = FALSE]
dg40_cn = dg40$copynumber[, dg40$stats[classifier_prediction == "good", sample], with = FALSE]
# Add prefix to generate unique colnames
setnames(dg20_cn, paste0("dg20_", colnames(dg20_cn)))
setnames(dg21_cn, paste0("dg21_", colnames(dg21_cn)))
setnames(dg22_cn, paste0("dg22_", colnames(dg22_cn)))
setnames(dg33_cn, paste0("dg33_", colnames(dg33_cn)))
setnames(dg39_cn, paste0("dg39_", colnames(dg39_cn)))
setnames(dg40_cn, paste0("dg40_", colnames(dg40_cn)))
# Combine libraries with same cell type
donor1_neun = cbind(dg20_cn, dg21_cn)
donor1_skmusc = dg22_cn
donor2_neun = cbind(dg33_cn, dg39_cn)
donor2_skmusc = dg40_cn
# Make annotation for brain cells
donor1_annot = data.table(sample = colnames(donor1_neun),
variable = "celltype",
value = c(rep("NeuN+", ncol(dg20_cn)), rep("NeuN-", ncol(dg21_cn))))
donor2_annot = data.table(sample = colnames(donor2_neun),
variable = "celltype",
value = c(rep("NeuN", ncol(dg33_cn)), rep("NeuN+", ncol(dg39_cn))))
# Plot heatmaps for donor 1
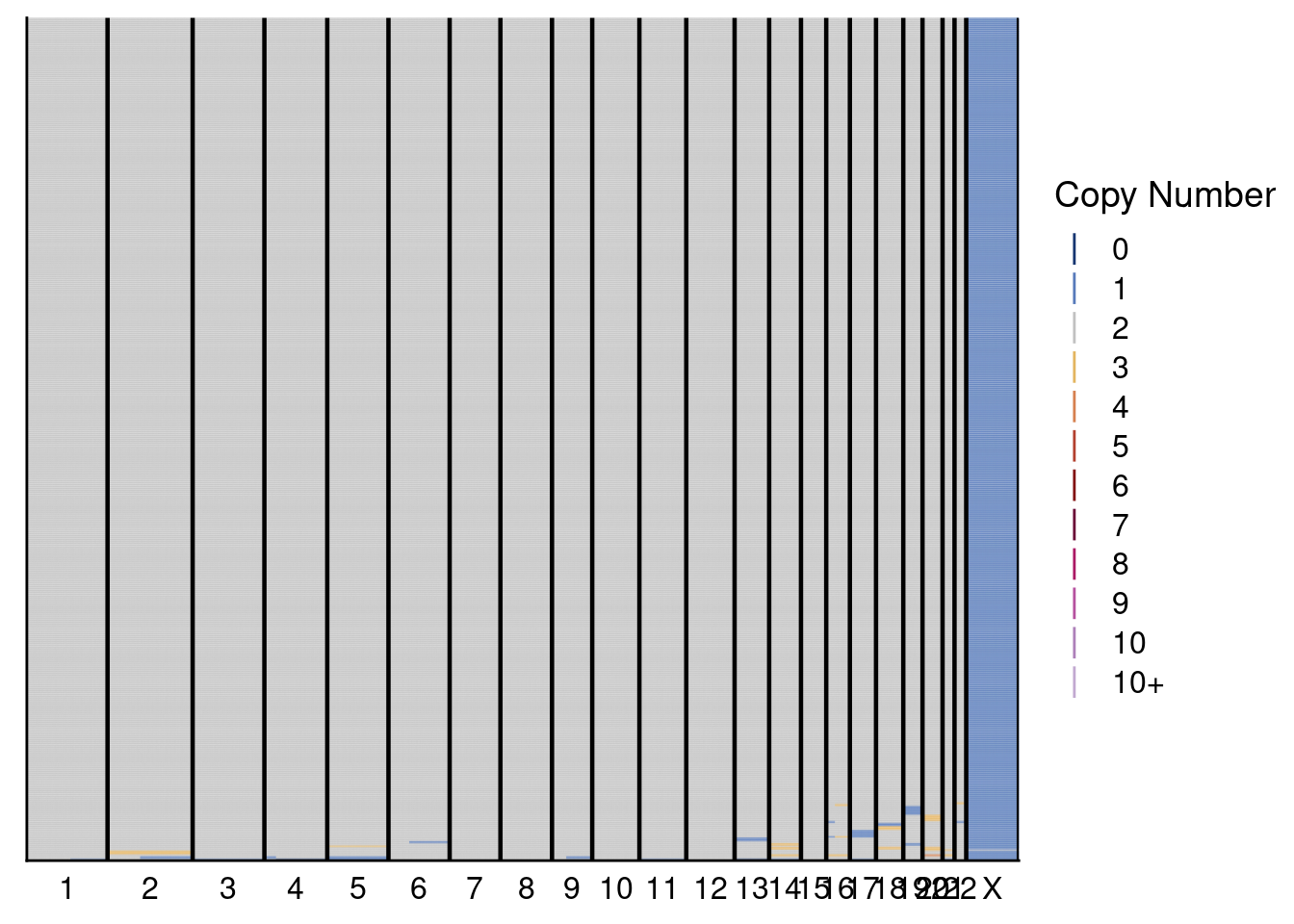
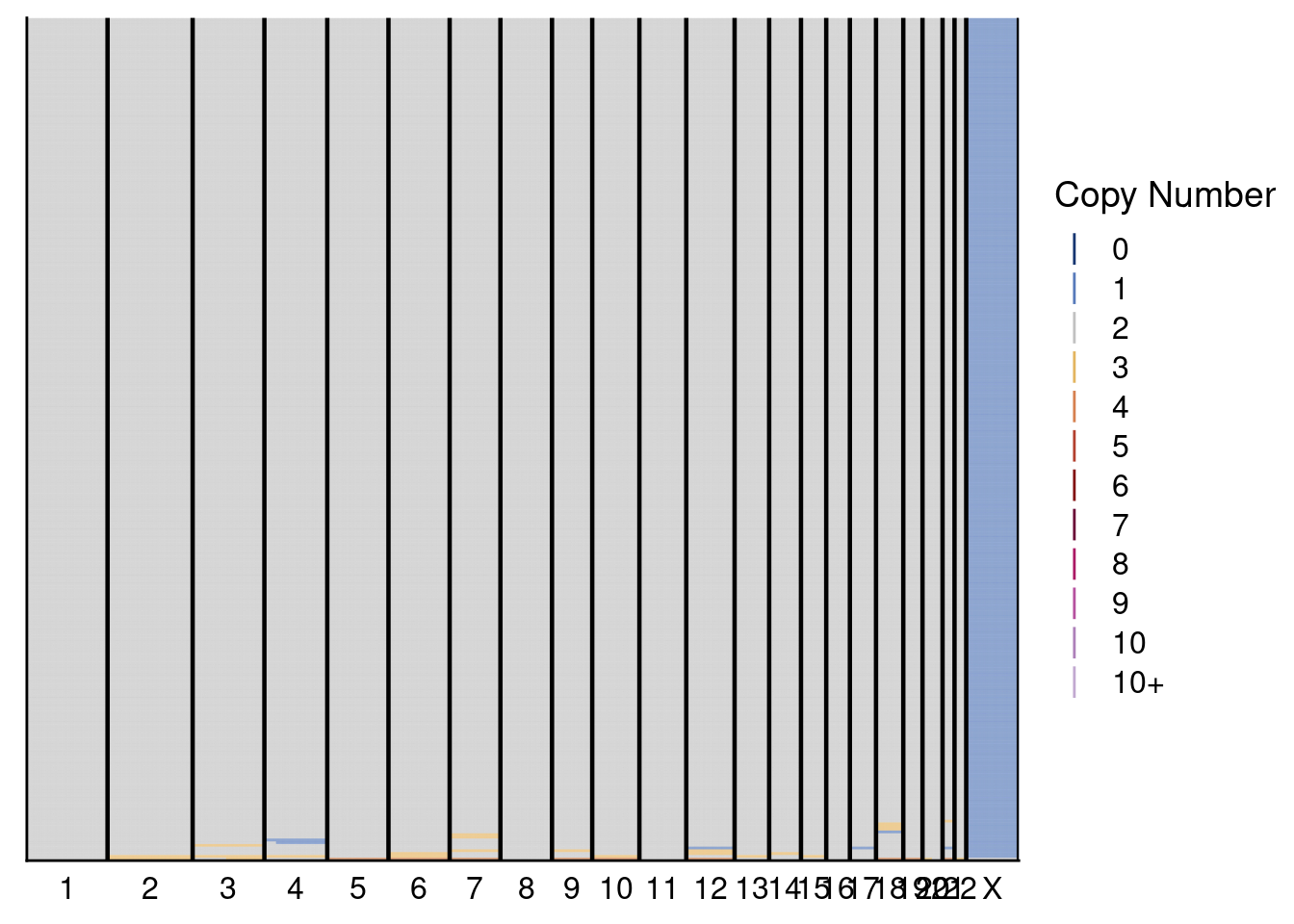
plotHeatmap(donor1_neun, dg20$bins, annotation = donor1_annot, dendrogram = FALSE)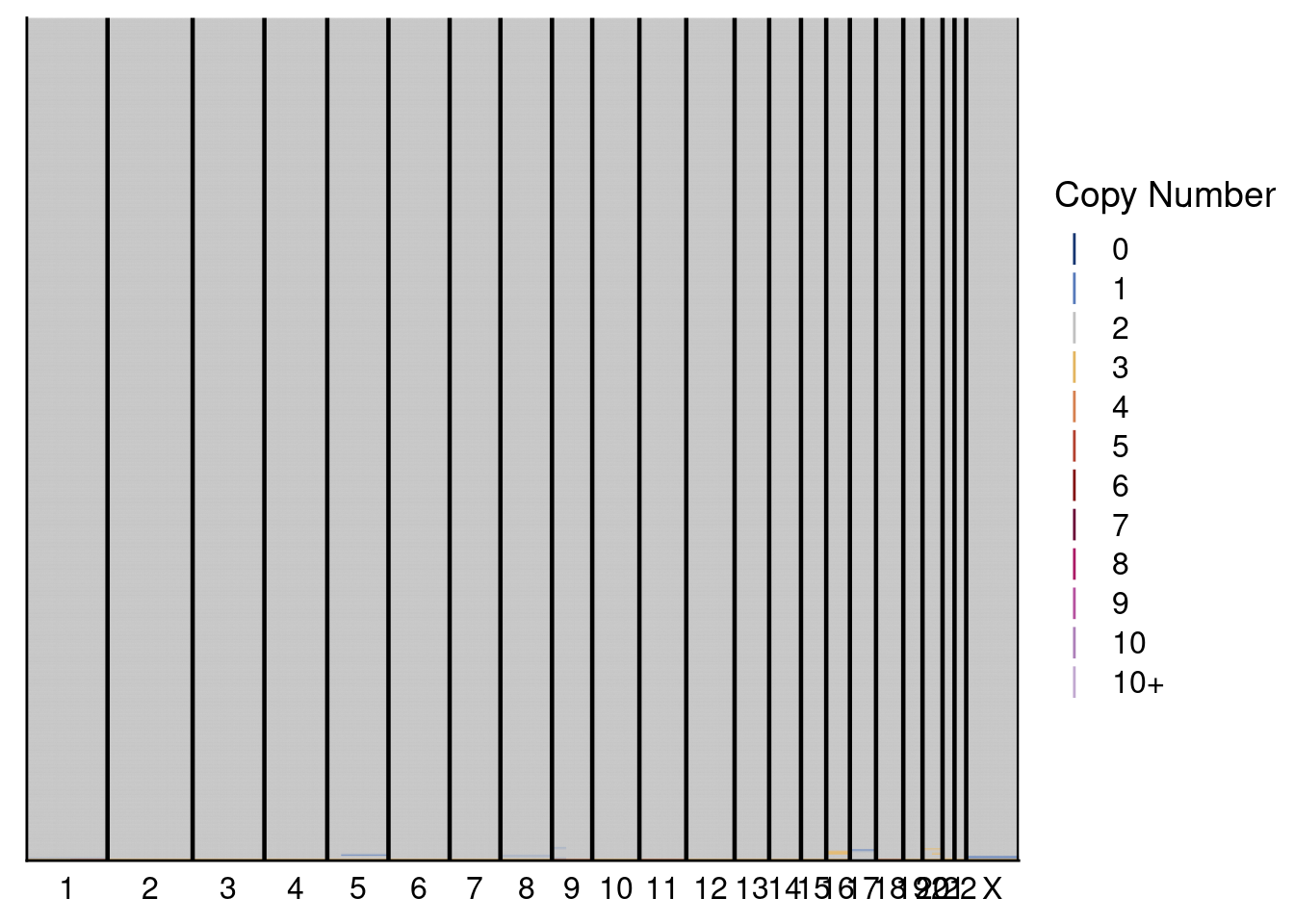
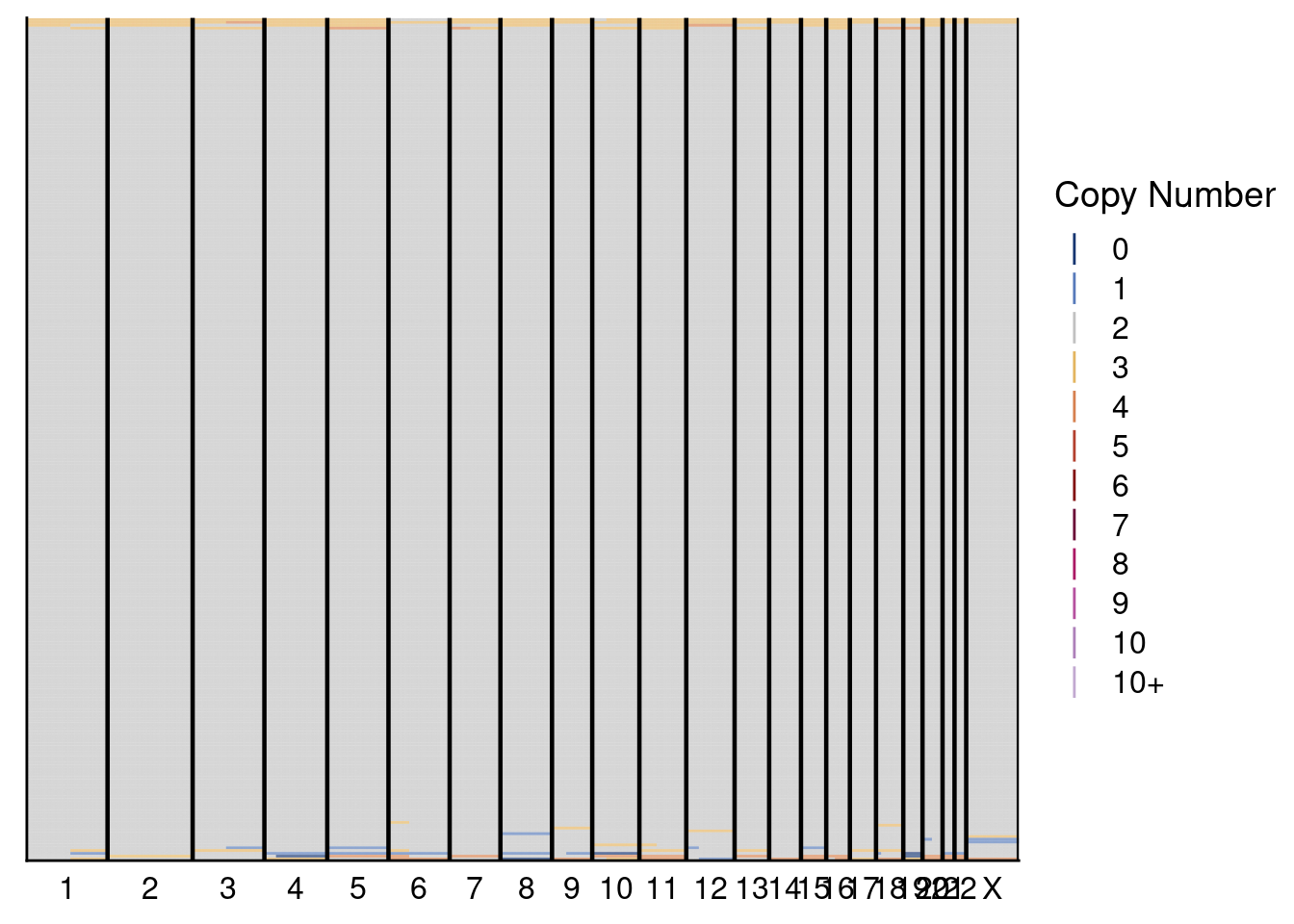
# Plot heatmaps for donor 2
plotHeatmap(donor2_neun, dg20$bins, annotation = donor2_annot, dendrogram = FALSE)